Amazing stuff! Solving the enigma of enzymes!
"Stanford researchers have illuminated how enzymes are able speed up life-sustaining biochemical reactions so dramatically. Their discoveries could impact fields ranging from basic science to drug discovery. ...
Using a series of more than 1,000 X-ray snapshots of the shapeshifting of enzymes in action, researchers at Stanford University have illuminated one of the great mysteries of life – how enzymes are able speed up life-sustaining biochemical reactions so dramatically. ...
“When I say enzymes speed up reactions, I mean as in a trillion-trillion times faster for some reactions,” ...
As a result, biochemists don’t have a basic understanding and, therefore, have been unable to predict enzyme rates or design new enzymes as well as nature does, an ability that would be impactful across industry and medicine. ...
that enzymes are not a single structure. Instead, they focus on what they term “ensembles,” showing how enzymes move between different physical states – or conformational ensembles – during catalysis. ...
Exploring these ensembles and comparing reaction states on enzymes to states of uncatalyzed enzymes in pure water, ... broke down enzyme catalysis to the individual energetic contributions at the precise place where enzyme and target molecule meet during a reaction, known as the active site, to understand how they work chemically and physically to speed up reactions. ...
“Nature has evolved these mechanisms independently in multiple enzyme families – this is not an isolated feature, but catalytic mechanisms that have been discovered multiple times by nature through evolution. This means we may be able to copy nature and use this and other features to design and build new enzymes,” ..."
From the editor's summary and abstract:
"Editor’s summary
Three-dimensional structures of enzymes have provided important insights into the chemical mechanisms through which these catalysts achieve their incredible rate accelerations. Building on decades of enzymology and structural biology, Du et al. analyzed more than 1000 structures of serine proteases to develop a comprehensive and quantitative framework for the origins of rate acceleration in this enzyme family. The authors highlight precise positioning of catalytic residues, ground state destabilization, and motion that is flexible but restricted along a productive path as key factors in rate acceleration by serine proteases. ...
Structured Abstract
INTRODUCTION
Enzymes are central to all biological functions. A century of enzymology has established chemical mechanisms and roles of active-site residues and cofactors for almost all known enzymes.
Yet, we still lack the ability to account for the enormous rate enhancements provided by enzymes.
Although serine proteases are often used as examples in biochemistry textbooks to illustrate enzyme catalysis, it remains unclear how their active sites accelerate the peptide bond cleavage reaction.
The serine nucleophile is a hydroxyl group that is similar to water in reactivity; the oxyanion hole typically consists of amide groups, which are also similar to water as hydrogen bond donors. The general base in the catalytic triad does facilitate proton transfer but not nearly enough to account for serine protease catalysis. A deepened understanding of how such enzymes work requires an approach that can identify catalytic features, uncover the physical basis of their function, and quantify their catalytic contributions.
RATIONALE
Reaction rates—and catalysis—are determined by the probability of achieving the reaction’s transition state, and these probabilities are defined by free energies. Conformational ensembles encode states and their probabilities, thereby providing a means to access the free energies that define catalysis.
We therefore moved beyond traditional structure-function relationships to develop an ensemble-function approach that identifies catalytic features and quantifies the catalytic contribution from each individual feature.
We applied this approach to serine proteases to address the following questions:
(i) What are the molecular changes between the enzymatic reaction states?
(ii) How does the enzymatic reaction differ from that in solution? and
(iii) What are the energetic consequences of these differences?
RESULTS
We brought together >1000 x-ray crystallographic structures from 17 serine proteases to build pseudo-ensembles, and we obtained ensemble information for the uncatalyzed solution reaction from small-molecule structures and computation. Pseudo-ensembles for enzymes bound to ground state analogs, transition state analogs, or no substrates revealed an altered energy landscape for the enzymatic reaction, where shorter and more efficient reaction paths are favored relative to those in solution.
The enzymes position reactants within a short distance and use a side-chain rotation of the catalytic serine to facilitate reaction, removing much of the translational motion needed in solution and increasing the reaction probability. The enzymatic reaction is further accelerated by multiple interactions that are destabilized in the ground state and relieved in the transition state.
We quantified catalytic contributions from these features using knowledge-based energy functions to provide a minimal model that accounts for serine protease catalysis within error. The same catalytic features are commonly used by enzymes from distinct structural folds to catalyze reactions that involve nucleophilic addition on carbonyl compounds, suggesting convergent evolution under the same mechanistic constraints and high evolvability of the identified strategies.
CONCLUSION
Ensemble-function analyses identified precise catalytic features and established an experimentally based, quantitative model for serine protease catalysis. This model is grounded in the most basic concepts of chemistry and physics—torsion angles, van der Waals interactions, hydrogen bonds, and conformational entropy—and thus is accessible to researchers and students alike. The ensemble-function approach connects these fundamental physicochemical properties to the emergent functions of macromolecules, a connection that is needed to understand enzyme catalysis and still more complex protein functions, such as allostery and molecular machines."
Conformational ensembles reveal the origins of serine protease catalysis (no public access, but here is link to the preprint)
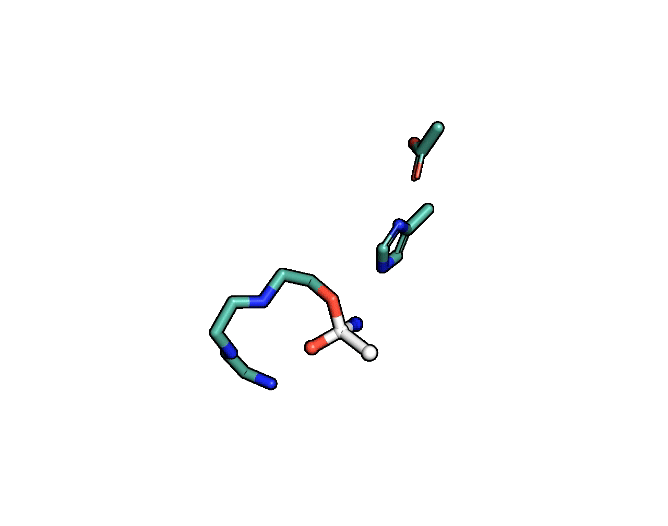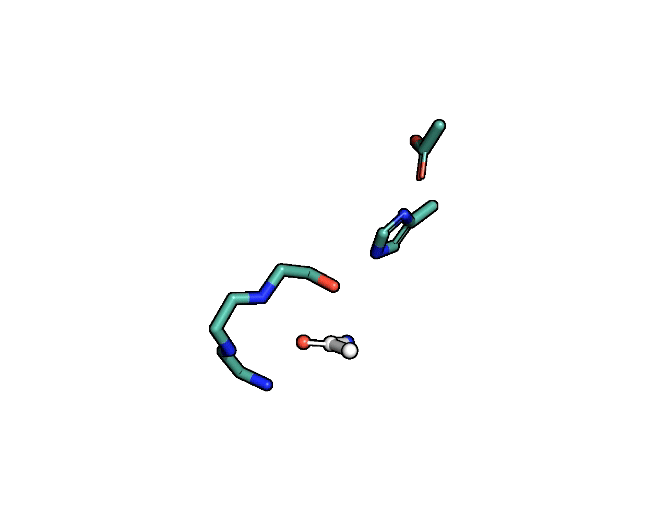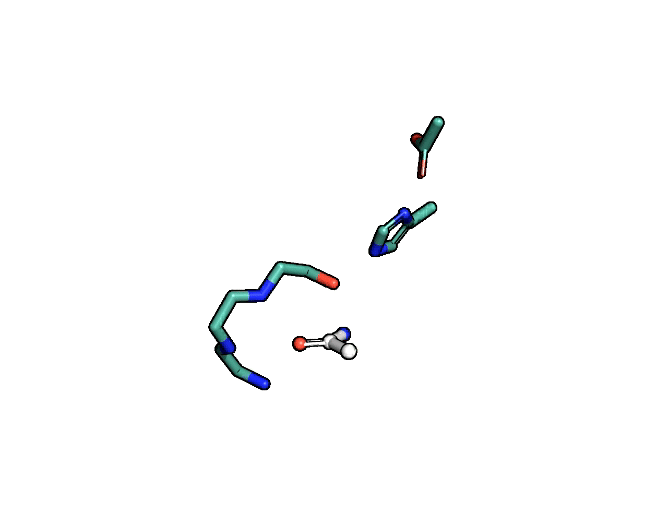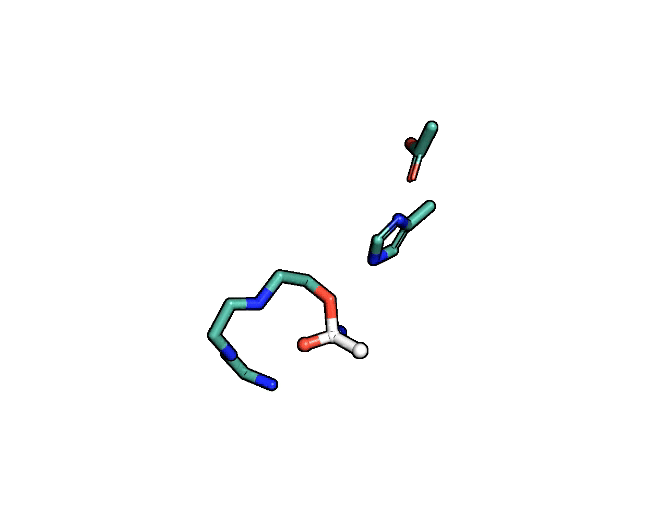
Animation showing the transition state of a serine protease – an example of how enzymes are always in motion.
A close-up showing the serine protease reaction at the active site, place where enzyme and target molecule meet during a reaction.
The ensemble-function approach provides a quantitative catalytic model for serine proteases and identifies repeatedly evolved catalytic strategies.

No comments:
Post a Comment