Good news!
"Scientists ... have developed a novel method that uses artificial intelligence (AI) and advanced imaging techniques to more accurately and efficiently identify therapeutic antibodies to treat infectious diseases.
The breakthrough method ... reduces the time needed to identify protective antibodies from weeks to under a day—while offering a scalable approach that minimizes data bottlenecks and accelerates research. This advancement could transform how researchers develop treatments for influenza, HIV and other infectious diseases, particularly during health emergencies where rapid response is critical.
“This represents a paradigm shift in how we discover antibodies,” ... “By harnessing AI to analyze the structural details of immune responses, we can now identify the most promising therapeutic candidates in mere hours, with better success rates than traditional methods. This could be game-changing for pandemic preparedness and therapeutic development.” ..."
From the abstract:
"Antibodies are crucial therapeutics, comprising a substantial portion of approved drugs due to their safety and clinical efficacy.
Traditional antibody discovery methods are labor-intensive, limiting scalability and high-throughput analysis.
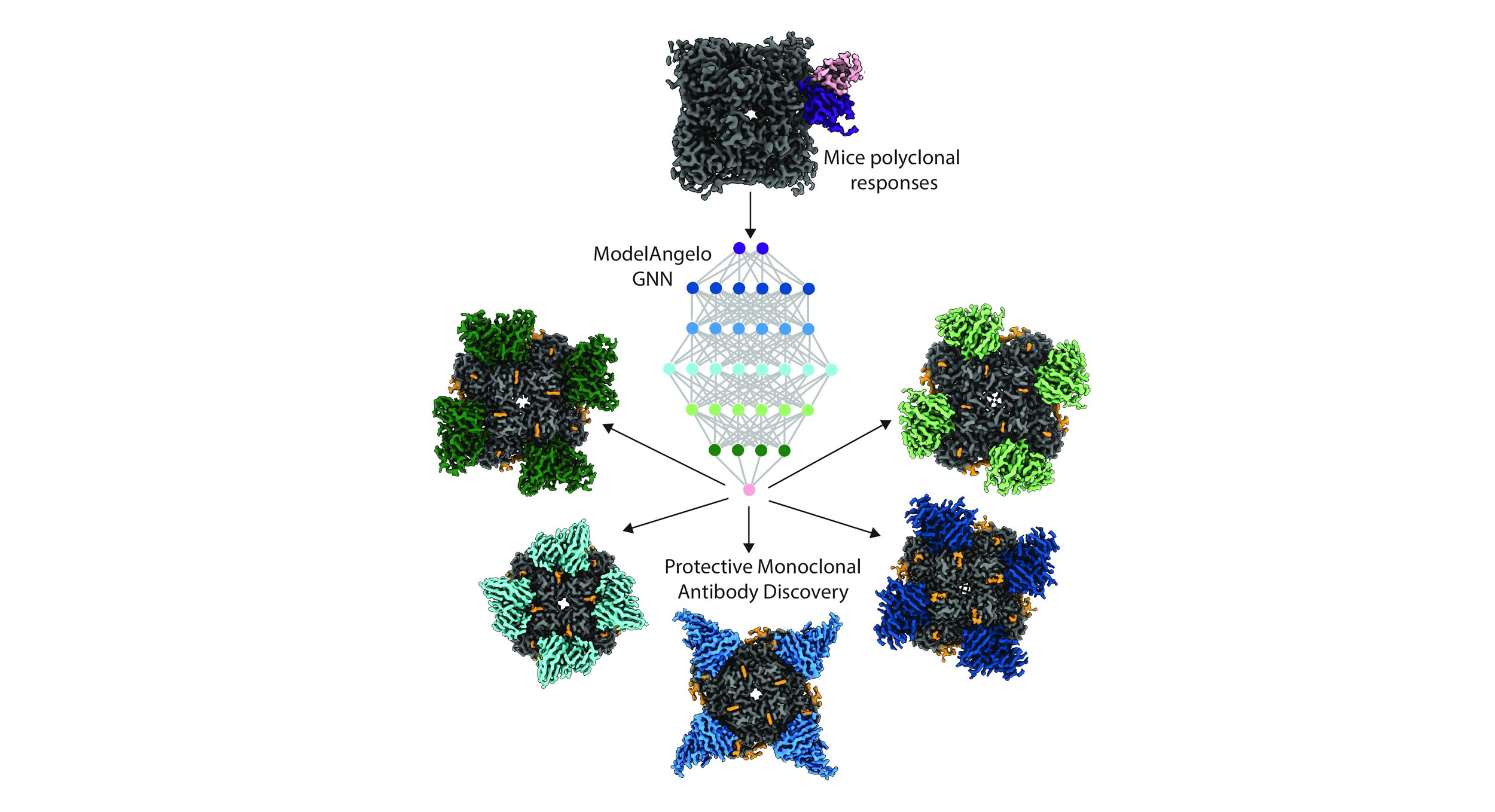
Here, we improved upon our streamlined approach combining structural analysis and bioinformatics to infer heavy and light chain sequences from cryo-EM (cryo–electron microscopy) maps of serum-derived polyclonal antibodies (pAbs) bound to antigens.
Using ModelAngelo, an automated structure-building tool, we accelerated pAb sequence determination and identified sequence matches in B cell repertoires via ModelAngelo-derived hidden Markov models (HMMs) associated with pAb structures.
Benchmarking against results from a nonhuman primate HIV vaccine trial, our pipeline reduced analysis time from weeks to under a day with higher precision. Validation with murine immune sera from influenza vaccination revealed multiple protective antibodies. This workflow enhances antibody discovery, enabling faster, more accurate mapping of polyclonal responses with broad applications in vaccine development and therapeutic antibody discovery."
Functional and epitope specific monoclonal antibody discovery directly from immune sera using cryo-EM (open access)
Scripps Research scientists used a graphical neural network-based structure building tool, ModelAngelo, to discover monoclonal antibodies (bottom) from polyclonal antibody responses produced after mouse vaccination (top).
Fig. 1. MA integrated STS workflow and benchmarking.

No comments:
Post a Comment